扩展功能
文章信息
- 梁昊, 刘红莹, 尤元海, 顾一心, 张艾煜, 王敏, 何利华, 孟凡亮, 张建中, 张茂俊
- LIANG Hao, LIU Hong-ying, YOU Yuan-hai, Gu Yi-xin, ZHANG Ai-yu, WANG Min, HE Li-hua, MENG Fan-liang, ZHANG Jian-zhong, ZHANG Mao-jun
- 利用基因芯片技术分析空肠弯曲菌的遗传特征
- Genetic analysis with DNA microarray for Campylobacter jejuni isolated in China
- 疾病监测, 2016, 31(2): 159-165
- Disease Surveillance, 2016, 31(2): 159-165
- 10.3784/j.issn.1003-9961.2016.02.017
-
文章历史
- 收稿日期: 2015-09-30
自2000年2月第一株空肠弯曲菌(Campylobacter jejuni)NCTC11168的全基因组序列完成图发布以来,陆续有多株空肠弯曲菌完成了全基因组测序。对不同菌株的序列比较分析显示空肠弯曲菌遗传变异度较大[1, 2, 3, 4, 5, 6, 7, 8]。这些全基因组序列为设计基因芯片探针提供了依据。根据基因组的任何一部分序列设计寡核酸探针,使研究者可利用高通量的基因芯片在基因组水平上研究大规模菌株的遗传特征[9, 10]。
目前,中国空肠弯曲菌菌株尤其是格林-巴利综合征(Guillain-Barre Snydrome,GBS)相关菌株的遗传特征的报道不多,为获得空肠弯曲菌尤其是GBS相关菌株的遗传特征,本研究在前期将5株已获得全基因组序列的国外菌株与自主完成全基因组测序的中国GBS暴发相关菌株ICDCCJ07001比对分析的基础上设计了包括所有ICDCCJ07001染色体基因、2个与空肠弯曲菌耐药和致病相关的质粒基因及5株国外菌株相对ICDCCJ07001特异基因的探针。同时为了获得菌株脂寡糖(lipooligosaccharides,LOS)合成基因簇及血清分型相关基因簇的特征,探索利用DNA序列分析代替传统血清分型方法的可行性,芯片设计了16种LOS合成相关基因簇及7种血清型相关的荚膜多糖(polysaccharide capsule,CAP)合成相关基因簇的探针,对包括9株GBS相关菌株在内的不同宿主来源的27株空肠弯曲菌菌株进行研究。
1 材料与方法 1.1 材料GBS相关空肠弯曲菌9株,其中分离GBS暴发来源1株,分离GBS散发病例8株,腹泻病例来源10株,鸡粪来源6株,食物来源2株。所有研究菌株在含5%脱纤维羊血的Kamali选择性培养基(Oxoid CM0935B)中于42 ℃微需氧条件下(5% O2、10% CO2和85% N2)培养48 h,利用QIAGEN DNA提取试剂盒(Qiagen)按说明书提取所有菌株的基因组DNA。
1.2 方法 1.2.1 芯片设计方案采用CombiMatrix tiling CustomArrayTM 90K芯片,在前期比较基因组分析的基础上,设计芯片并合成探针。芯片上涵盖探针序列包括5部分:(1)基因组完成测序菌株ICDCCJ07001中的所有编码蛋白产物的序列(Coding sequence,CDS)。(2)空肠弯曲菌常见两种质粒ICDCCJ07001_pTet及81-176_pVir所有CDS。(3)5株不同测序菌株与菌株ICDCCJ07001相比特异基因的CDS。(4)与空肠弯曲菌CAP合成相关的基因CDS。(5) LOS合成相关的基因CDS,具体探针涉及基因内容见表 1。
| CDS来源 | 菌株来源 | GenBank 序列号 | CDS个数 | |
| 1 | ICDCCJ07001所有CDSs | ICDCCJ07001 | NC_014802.1 | 1634 |
| 2 | 质粒相关基因 | ICDCCJ07001_pTet | NC_014801.1 | 37 |
| 81-176_pVir | NC_008770 | 53 | ||
| 3 | 菌株特异基因 | 269.97 | NC_009707 | 542 |
| NCTC11168 | NC_002163 | 280 | ||
| 81-176 | NC_008787 | 243 | ||
| 81116 | NC_009839 | 249 | ||
| RM1221 | NC_003912 | 469 | ||
| 4 | 荚膜多糖相关基因簇 | 260.94 | NZ_AANK01000002 | 35 |
| CG8486 Scon20 | NZ_AASY01000009 | 8 | ||
| CG8486 Scon22 | NZ_AASY01000001 | 16 | ||
| CG8421 | ABGQ01000005 | 25 | ||
| 5 | 脂寡糖相关基因簇 | QYT LOS gene cluster | DQ864651 | 13 |
| ZX LOS gene locus | DQ535891 | 19 | ||
| LC LOS gene locus | DQ535892 | 19 | ||
| putative heptosyltransferase I gene… | AY422196 | 14 | ||
| RM2095 LOS biosynthesis locus | AY816330 | 6 | ||
| GB4 LOS gene locus | AY943308 | 18 | ||
| GC149 LOS gene locus | AY962325 | 13 | ||
| ATCC43437 LOS biosynthesis locus | AY436358 | 9 | ||
| RM1047 LOS biosynthesis locus | AY800272 | 16 | ||
| LIO87 LOS biosynthesis locus | AF400669 | 10 | ||
| RM1850 class I LOS biosynthesis gene locus | EU404107 | 12 | ||
| RM1508 class J LOS biosynthesis gene locus | EU404104 | 11 | ||
| RM1861 LOS biosynthesis locus | EU410350 | 9 | ||
| RM3435 class L LOS biosynthesis gene locus | EU404111 | 6 | ||
| RM3437 class Q LOS biosynthesis gene locus | EU404112 | 9 | ||
| RM3419 class S LOS biosynthesis gene locus | EU404110 | 9 | ||
| RM3423 class O LOS biosynthesis gene locus | EF143352 | 16 | ||
| RM1503 class M LOS biosynthesis gene locus | EF140720 | 10 |
芯片杂交方法参照You等[11]的论著,将浓缩的Cy5标记的DNA悬浮在90 μl含50%甲酰胺、3×SSC、0.1% SDS、5×登哈特溶液的杂交液和0.1 mg/ml的鲑鱼精子DNA中[12]。被标记的DNA在85 ℃、30 min条件下变性,然后快速置于冰上1 min,放入分子杂交仪上于65 ℃杂交10 min。将杂交好的芯片用SSPET和PBS洗脱液吹洗后,在Gene Pix4000仪器上扫描,图像保存。
1.2.3 数据分析扫描后的芯片图片导入Microarray Image 5.9.3软件,以列表形式导出每个点的荧光信号强度值;将芯片上的质控点及空白点的信号值作为背景值,统计分析后得到所有菌株杂交后的背景信号值均 < 250,若一个基因中≥80%探针的信号值≥500,则该基因为参比基因组/簇所含有的基因,不符合此条件的基因为变异或缺失的基因;按照上述定义分析27株菌的每个基因的状态,用Mev v4.0软件进行分层聚类分析。
2 结果 2.1 4组空肠弯曲菌CDS的分布特征ICDCCJ07001、 269.97、NCTC11168、81-176、81-116和RM1221 6个菌株的基因组中共包含3384种CDS,将27株菌的CDS与这6株菌的CDS进行比较分析,所有33株菌共同包含的CDS为589个。将这6株全基因组测序的菌株的有CDS建库,与库中的CDS相比,27株菌中共同包含的CDS为929个(图 1)。GBS组、腹泻组、鸡粪组、食物组分别与库中比对上了1301、1191、1308和1763个CDS。

|
| 注:红色部分与库中比对上的且4组共有的CDS,蓝色为与库中比对上但4组各自特异CDS。 图 1 4组空肠弯曲菌CDSs与6株全基因组测序菌株CDS库的比较 Figure 1 Comparison of CDSs of four groups and CDS Library of 6 whole genome sequencing strains of C. jejuni |
将27株菌与ICDCCJ07001染色体基因组杂交结果进行比较并进行聚类分析,找到4个变最显著的区域分别是1-CMLP1相关基因、2-LOS合成相关基因、3-鞭毛修饰相关基因、4-CAP合成相关基因。聚类分析结果未发现明显的宿主特征性。各区域包含的CDS名称及其功能见表 2。
| 变异区域编号 | 基因 | 功能 |
| 1 | ICDCCJ07001_659-699 | Campylobacter Mu-like phage (CMLP1) |
| 2 | ICDCCJ07001_1091-1110 | lipooligosacchride(LOS) |
| 3 | ICDCCJ07001_1249-1274 | flagellin modification(FM) |
| 4 | ICDCCJ07001_1344-1382 | capsular polysaccharide(CAP) |
ICDCCJ07002几乎含有ICDCCJ07001_pTet的所有CDS,其余26株菌中有18株菌含有部分pTet的CDS,11株菌含有35%以上的基因,7株菌只含有CDS ICDCCJ07001_pTet000007;有6个ICDCCJ07001_pTet的CDS在10株以上的菌株的质粒中,分别是ICDCCJ07001_pTet000004、ICDCCJ07001_pTet000010、ICDCCJ07001_pTet000020、ICDCCJ07001_pTet000021、ICDCCJ07001_pTet000028、ICDCCJ07001_pTet000034。27株菌中有10株含有CJJ81176_pVir0018、 CJJ81176_pVir0041、CJJ81176_pVir0042、CJJ81176_pVir0048 这4个81-176_pVir CDS中的1~2个;聚类结果未发现宿主特征性,见图 2。
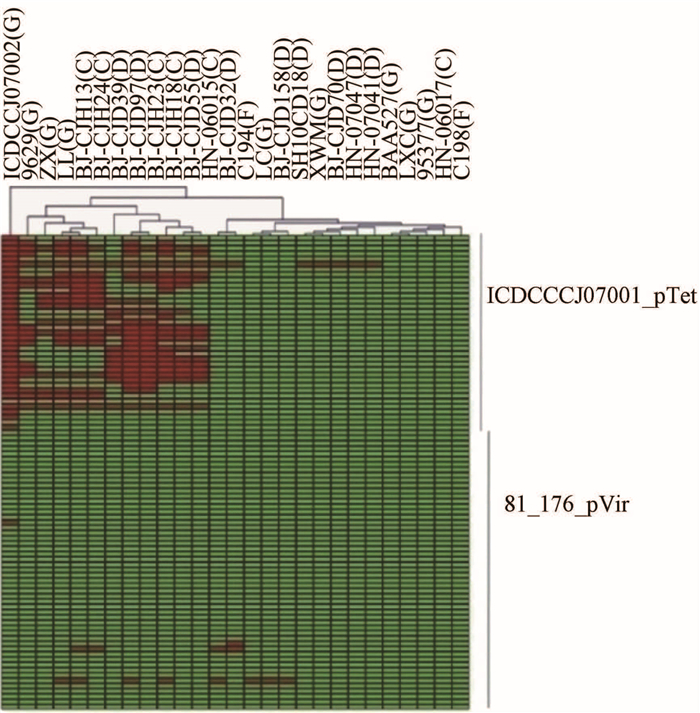
|
| 注:G-GBS组,D-腹泻组,C-鸡粪组,F-食物组。图中每列代表1株菌,每行代表1个基因。红色代表与参比基因簇共有的CDS,绿色代表变异或缺失CDS,下同。 图 2 27株菌与2个质粒CDSs比较及聚类分析 Figure 2 Comparison and cluster analysis of 27 strains and two plasmid CDSs |
该部分结果为27株菌与5株国外菌株相对ICDCCJ07001特异的CDS的检测。结果表明,所有27株菌几乎都不含有269.97的特异CDS,部分含有NCTC11168、81-176、81-116、RM1221的特异CDS,在RM1221的特异CDS中,有4个明显变异的区域:空肠弯曲菌Mu样噬菌体基因簇(Campylobacter Mu-like phage,CMLP)1~4中[13, 14, 15, 16, 17, 18, 19],分别是CMLP1、CMLP2、CMLP3、CMLP4。通过对27株菌中4个CMLP相关基因簇的基因与RM1221进行同源性比较发现,同源率≤30%的占绝大部分,见图 3。
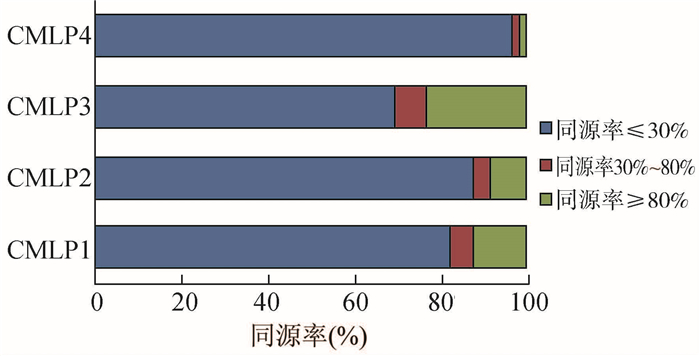
|
| 图 3 27株菌中4个CMLP基因簇与RM1221同源率的比较 Figure 3 Homology comparison of 4 CMLP gene clusters of 27 strains and the strain of RM1221 |
如图 4所示,芯片上包括了16种LOS类型的探针,若一个菌株在一个LOS类型相关的基因簇中有1个以下的基因是变异或缺失基因的话,则认为此LOS型为这株菌的LOS类型。可以确定LOS类型的菌株共14株,占总菌株的52%;在导致GBS的9株菌中,有7株确定了LOS型,其中5株属于A型。
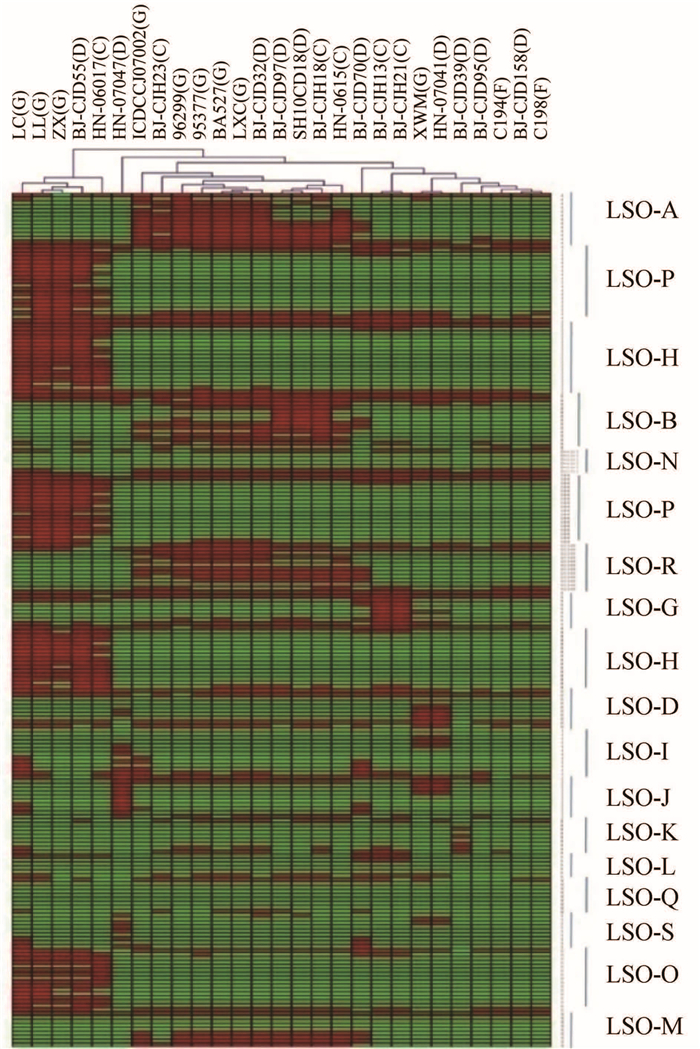
|
| 图 4 27株菌LOS型相关基因比较及聚类分析结果 Figure 4 Comparison and clustering analysis of LOS genotypes of 27 strains |
本次研究设计了7种血清型的血清型的基因芯片(HS41、HS4、HS3、HS2、HS23/36、HS6和HS53)。在27株菌中检测出1株HS41血清型菌株(HN-06015),2株HS2血清型菌株(96299和BJ-CJH18),2株HS53血清型菌株(XWM和BJ-CJD95),未检出其他血清型,具体结果见图 5。
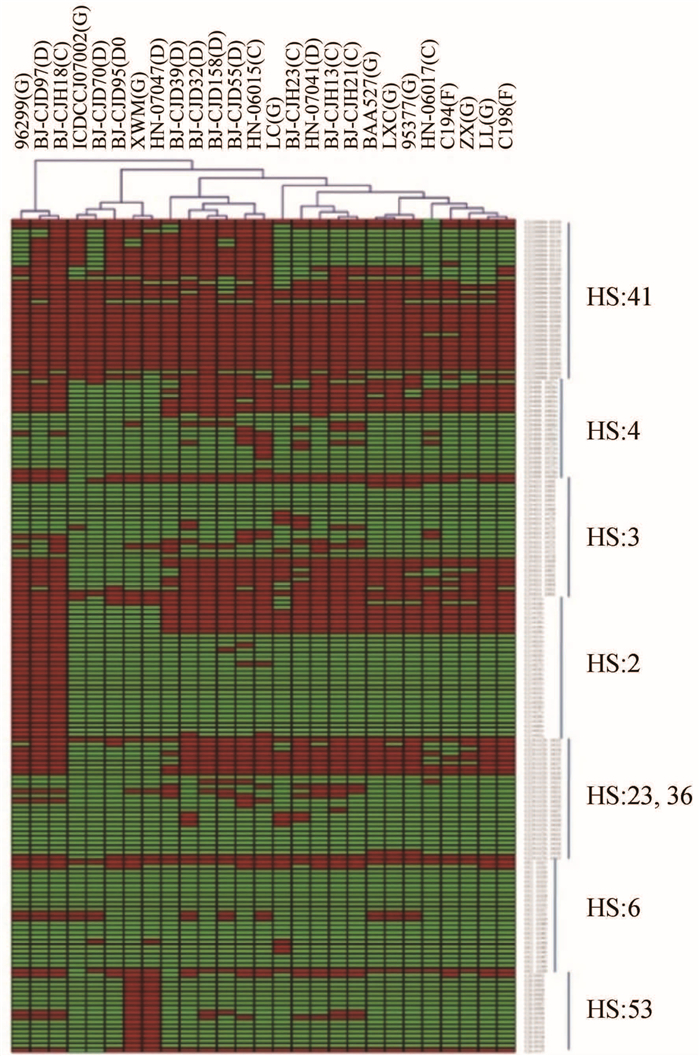
|
| 图 5 27株菌血清型相关基因比较聚类分析 Figure 5 Comparison and clustering analysis of serotypes of 27 strains |
基于全基因组测序的基因组比较分析能够提供最为准确详细的信息,但是由于全基因组测序耗时过长,成本过高,而基于芯片技术的比较基因组杂交(aCGH)成为进行大量菌株的遗传特征分析的有效方法。利用基因芯片进行空肠弯曲菌的遗传特征及流行病学分析已有不少先例[13, 14, 15, 16, 17, 18, 19],然而这些研究中设计的探针覆盖范围小,导致大量信息无法获得。本研究所设计的芯片涵盖了6株全基因组测序菌株ICDCCJ07001、269.97、NCTC11168、81-176、81-116和RM1221的所有CDS,空肠弯曲菌中常见的两个与致病相关的质粒ICDCCJ07001_pTet和81-176_pVir,16种LOS型相关基因以及7种血清型相关的基因。而目前研究报道一些特异菌型的空肠弯曲菌,如特异LOS型及血清型,与GBS高度相关[20, 21, 22, 23, 24, 25],因此特异菌型及遗传特征的病原分析对于空肠弯曲菌感染导致GBS的诊断具有重要价值。
本次研究对27株中国菌株与6株模式菌株的全基因组进行了比对,通过对这些菌株CDS的分布分析,结果显示,研究所涉及的我国27株菌株与6株模式菌株的核心基因组共589个CDS[26],这些基因多参与代谢及合成等进程,对于这部分基因笔者均进行了PCR验证,证明所有菌株都含有这些CDS,表明本研究方法结果准确。各菌株的CDS总数目在1598~2254之间,核心基因大约占到总基因组基因数量的26.1%~36.9%。269.97、NCTC11168、81-176、81-116和RM1221是已经完成了全基因组测序5株空肠弯曲菌国外菌株,本课题组在之前的研究中将自主完成测序的中国菌株ICDCCJ07001与这5株菌进行了全基因组比较[4],获得了5株菌相对于ICDCCJ07001的特异CDS,并在芯片上设计了针对这些菌株特异CDS的探针。269.97属于空肠弯曲菌德莱亚种,该亚种与空肠亚种相比具有其独特的遗传特征[27]。而本研究所选择的27株菌均为空肠弯曲菌空肠亚种,芯片结果看到269.97的特异CDS在所有27株中国菌株均缺失或高度变异,与预期结果一致;NCTC11168、81-176、81-116均分离自胃肠炎患者的粪便,属空肠亚种,可以看到这3株菌的CDS在中国菌株中仍有存在。以上结果表明,在地域方面,我国菌株与国外菌株相比存在大量的同源基因,在弯曲菌防控方面,应更多开展国际上合作。
RM1221是一株分离自鸡表皮的空肠弯曲菌,它与其他菌株最显著的不同在于其基因组中含有4个空肠弯曲菌整合元件CMLP1、CMLP2、CMLP3和CMLP4[28],27株中国菌株中,这4个空肠弯曲菌整合元件相关基因绝大多数与RM1221同源率在30%以下,说明27株中国菌株大多不含CMLP相关基因。
ICDCCJ07001_pTet是一个四环素耐药质粒,共37个基因[4],ICDCCJ07002几乎含有该质粒的全部基因,其余26株菌中有18株菌含有部分pTet基因,其中11株菌含有35%以上该质粒基因,但这18株菌中都含有与四环素耐药先关的基因ICDCCJ07001_pTet000007基因。81-176_pVir是与细菌粘附侵袭相关的毒性质粒[29],大小为 37 468 bp,共53个基因[30],pVir质粒在空肠弯曲菌中的携带率较低,之前有加拿大和挪威的研究显示在104和146株空肠弯曲菌中,分别只有17.0%和3.4%的菌株携带该质粒。针对笔者菌株的81-176_pVir检测中,只有10株菌携带该质粒中的1~2个CDS,表明笔者的菌株中不含有该质粒的基因。
LOS是空肠弯曲菌细胞壁的成分,因为其结构与人神经细胞膜上的神经节苷脂成分结构相似,空肠弯曲菌感染人体后可产生抗神经节苷脂的抗体,该抗体与神经节苷脂发生免疫反应从而导致GBS[31, 32]。目前已知的空肠弯曲菌的LOS类型共有19种:A~S[22, 33],芯片上包含了16种不同类型LOS合成相关的基因的探针,27株菌中可以明确LOS类型的14株,在GBS组的9株菌中,有7株确定了LOS型,其中LOS-A 5株,LOS-P 2株,LOS-H 1株,LXC、LL、ZX菌株之前通过测序获得了其LOS类型[34],与通过芯片得出的结果一致,说明芯片检测结果的可靠性;另外,9株GBS相关菌株中大部分LOS类型为A型,A型LOS合成相关基因中有能够编码唾液酸转移酶的基因,能够指导合成与神经节苷脂结构类似的LOS,因此具有A型结构LOS的菌株更容易导致GBS[35]。所以,本研究的方法对于确定空肠弯曲菌的LOS型别准确可信,可作为GBS高致病性菌株的实验室诊断方法。
以空肠弯曲菌热稳定性(HS)抗原进行分型的Penner分型分型方法可以将空肠弯曲菌分为上百个血清型[25],荚膜多糖(CAP)是决定不同Penner血清型的主要因素[36]。目前研究发现,与GBS高度相关的血清型有HS:41、HS:2、HS:4、HS:19,此外,在肠炎患者中普遍存在而在GBS患者中偶有报道的血清型有HS1、HS3、HS5、HS8、HS23、HS37、HS44、HS53等[23, 24, 25]。本研究所用芯片设计了7种与致病相关的血清型相关的CAP基因的探针,检测到了3种血清型(2株HS2血清型、1株HS41血清型和2株HS53血清型)的菌株。后期96299和XWM两株菌的基因组测序分析与基因芯片血清型检测的结果一致,另外3株菌均进行了血清型试剂盒的检测,与基因芯片结果一致,表明基因芯片检测血清型结果可信。
不同空肠弯曲菌的主要变异区位于表面结构相关的基因簇:LOS及CAP合成相关基因、鞭毛修饰(FM)相关基因,DNA限制/修饰相关基因以及4个CMLP基因簇。本次研究表明,基因芯片结果准确可信,可作为分析空肠弯曲菌遗传特征及确定其血清型和LOS类型手段,对诊断空肠弯曲菌导致的感染和GBS有重要价值。
| [1] | Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences[J]. Nature,2000,403(6770):665-668. |
| [2] | Gundogdu O, Bentley SD, Holden MT, et al. Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence[J]. BMC Genomics,2007,8(1):162. |
| [3] | Pearson BM, Gaskin DJH, Segers RPAM, et al. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828)[J]. J Bacteriol,2007,189(22):8402-8403. |
| [4] | Zhang MJ, He LH, Li Q, et al. Genomic characterization of the guillain-barre syndrome-associated Campylobacter jejuni ICDCCJ07001 Isolate[J]. PLoS One,2010,5(11):e15060. |
| [5] | Cooper KK, Cooper MA, Zuccolo A, et al. Complete genome sequence of Campylobacter jejuni strain S3[J]. J Bacteriol,2011,193(6):1491-1492. |
| [6] | Luo Y, Sahin O, Dai L, et al. Development of a loop-mediated isothermal amplification assay for rapid, sensitive and specific detection of a Campylobacter jejuni clone[J]. J Vet Med Sci,2012,74(5):591-596. |
| [7] | Revez J, Schott T, Rossi M, et al. Complete genome sequence of a variant of Campylobacter jejuni NCTC 11168[J]. J Bacteriol,2012,194(22):6298-6299. |
| [8] | Friis C, Wassenaar TM, Javed MA, et al. Genomic characterization of Campylobacter jejuni strain M1[J]. PLoS One,2010,5(8):e12253. |
| [9] | Barrett MT, Scheffer A, Ben-Dor A, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA[J]. Proc Natl Acad Sci USA,2004,101(51):17765-17770. |
| [10] | You YH, Zhang JZ. Data mining from microarray gene expression profile[J]. China Biotechnology,2009,29(10):87-91. (in Chinese) 尤元海,张建中. 基因表达谱芯片的数据挖掘[J]. 中国生物工程杂志,2009,29(10):87-91. |
| [11] | You YH, Fu CX, Zeng X, et al. A novel DNA microarray for rapid diagnosis of enteropathogenic bacteria in stool specimens of patients with diarrhea[J]. J Microbiol Methods,2008,75(3):566-571. |
| [12] | Denhardt DT. A membrane-filter technique for the detection of complementary DNA[J]. BiochemBiophys Res Commun,1966,23(5):641-646. |
| [13] | Dorrell N, Mangan JA, Laing KG, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity[J]. Genome Res,2001,11(10):1706-1715. |
| [14] | Leonard II EE, Takata T, Blaser MJ, et al. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates[J]. J Infect Dis,2003,187(4):691-694. |
| [15] | Parker CT, Quiñones B, Miller WG, et al. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni Strain RM1221[J]. J Clin Microbiol,2007,44(11):4125-4135. |
| [16] | Quiñones B, Guilhabert MR, Miller WG, et al. Comparative genomic analysis of clinical strains of Campylobacter jejuni from South Africa[J]. PLoS One,2008,3(4):e2015. |
| [17] | Rodin S, Andersson AF, Wirta V, et al. Performance of a 70-mer oligonucleotide microarray for genotyping of Campylobacter jejuni[J]. BMC Microbiol,2008,8(1):73. |
| [18] | Pearson BM, Pin C, Wright J, et al. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays[J]. FEBS Lett,2003,554(1/2):224-230. |
| [19] | Leonard II EE, Tompkins LS, Falkow S, et al. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barré syndrome and strains that cause enteritis by a DNA microarray[J]. Infect Immun,2004,72(2):1199-1203. |
| [20] | Godschalk PCR, Gilbert M, Jacobs BC, et al. Co-infection with two different Campylobacter jejuni strains in a patient with the Guillain-Barrésyndrome[J]. Microbes Infect,2006,8(1):248-253. |
| [21] | Gilbert M, Karwaski M F, Bernatchez S, et al. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide[J]. J Biol Chem,2002,277(1):327-337. |
| [22] | Parker CT, Horn ST, Gilbert M, et al. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources[J]. J Clin Microbiol,2005,43(6):2771-2781. |
| [23] | Takahashi M, Koga M, Yokoyama K, et al. Epidemiology of Campylobacter jejuni isolated from patients with Guillain-Barré and fisher syndromes in Japan[J]. J Clin Microbiol,2005,43(1):335-339. |
| [24] | Allos BM. Association between Campylobacter infection and Guillain-Barré syndrome[J]. J Infect Dis,1997,176 Suppl 2:S125-S128. |
| [25] | Penner JL, Hennessy JN. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens[J]. J Clin Microbiol,1980,12(6):732-737. |
| [26] | Dykhuizen DE, Green L. Recombination in Escherichia coli and the definition of biological species[J]. J Bacteriol,1991,173(22):7257-7268. |
| [27] | Parker CT, Miller WG, Horn ST, et al. Common genomic features of Campylobacter jejuni subsp. doylei strains distinguish them from C. jejuni subsp. jejuni[J]. BMC Microbiol,2007,7:50. |
| [28] | Fouts DE, Mongodin EF, Mandrell RE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species[J]. PLoS Biol,2005,3(1):72-85. |
| [29] | Bacon DJ, Alm RA, Burr DH, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176[J]. Infect Immun,2000,68(8):4384-4390. |
| [30] | Bacon DJ, Alm RA, Hu L, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176[J]. Infect Immun,2002,70(11):6242-6250. |
| [31] | Yuki N, Taki T, Inagaki F, et al. A bacterium lipopolysaccharide that elicits Guillain-Barré syndrome has a GM1 ganglioside-like structure[J]. J Exp Med,1993,178(5):1771-1775. |
| [32] | Aspinall GO, Fujimoto S, McDonald AG, et al. Lipopolysaccharides from Campylobacter jejuni associated with Guillain-Barré syndrome patients mimic human gangliosides in structure[J]. Infect Immun,1994,62(5):2122-2125. |
| [33] | Parker CT, Michel G, Yuki N, et al. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes:evidence of mosaic organizations[J]. J Bacteriol,2008,190(16):5681-5689. |
| [34] | Jiang H, Zhang MJ, Liu RC, et al. Characteristics of lipo-oligosaccharide loci of Campylobacter jejuni isolates associated with Guillain-BarréSyndrome from Hebei, China[J]. Int J Mol Sci,2010,11(3):1155-1161. |
| [35] | Gilbert M, Brisson JR, Karwaski MF, et al. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384 Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-MHz 1H and 13C NMR analysis[J]. J Biol Chem,2000,275(6):3896-3906. |
| [36] | Karlyshev AV, Champion OL, Churcher C, et al. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses[J]. Mol Microbiol,2005,55(1):90-103. |
 2016, Vol. 31
2016, Vol. 31


